Study Confirms Widespread Viability of Science Curriculum Developed at Pitt
For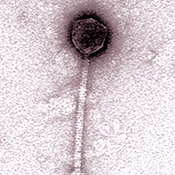 more than 30 years, Graham Hatfull has been happily studying and hunting down bacteriophages, viruses that infect bacteria and are greater in number than all other creatures on Earth combined. The Pitt biology professor is so passionate about “phages” (short for bacteriophages) that he’s now leading a movement to simultaneously study these viruses and improve science education on college campuses nationwide.
more than 30 years, Graham Hatfull has been happily studying and hunting down bacteriophages, viruses that infect bacteria and are greater in number than all other creatures on Earth combined. The Pitt biology professor is so passionate about “phages” (short for bacteriophages) that he’s now leading a movement to simultaneously study these viruses and improve science education on college campuses nationwide.
Hatfull’s success with developing a “phage-hunting” course—applicable to freshmen education at a range of institutions—is chronicled in a study published Feb. 4 in mBio, a peer-reviewed journal published by the American Society for Microbiology.
Hatfull, the Eberly Family Professor of Biotechnology in the University of Pittsburgh’s Department of Biological Sciences in the Kenneth P. Dietrich School of Arts and Sciences, began enlisting undergraduate Pitt students to aid him in his ongoing hunt for novel phages in 2001. The following year, he was named one of 20 Howard Hughes Medical Institute Professors tasked with developing creative, nationally applicable science education programs and putting them into practice. For the next several years, with the help of others, Hatfull created a course intended to infect freshman college students with the science bug. Called Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES for brevity’s sake), the course, formalized in 2008 at Pitt, now is being taught at 70-plus colleges and universities around the country.
To date, SEA-PHAGES students have found 3,000 new bacteriophages, all catalogued in Pitt’s Mycobacteriophage Database. The students get to name their own phages, which not only is fun but also gives them a sense of ownership over their contributions to phage research. Some of the students have also been credited as coauthors on 10 scientific journal articles. The course has proven to engage students deeply in STEM disciplines, and SEA-PHAGES “graduates” are more likely to continue in the sciences, one of the faster growing areas in today’s global economy.
date, SEA-PHAGES students have found 3,000 new bacteriophages, all catalogued in Pitt’s Mycobacteriophage Database. The students get to name their own phages, which not only is fun but also gives them a sense of ownership over their contributions to phage research. Some of the students have also been credited as coauthors on 10 scientific journal articles. The course has proven to engage students deeply in STEM disciplines, and SEA-PHAGES “graduates” are more likely to continue in the sciences, one of the faster growing areas in today’s global economy.
The study published in mBio analyzes the progress of more than 4,800 students who went through SEA-PHAGES over five years. The students took the course at a range of institutions—from research universities to colleges granting associate’s degrees. The bottom line is that SEA-PHAGES students considered themselves better educated than students who undertook intensive summer research programs and were more likely than their peers to stay involved in science, with some becoming undergraduate teaching assistants for the next group of phage-hunters.
Why? Because they got to do science, Hatfull says. SEA-PHAGES goes beyond a study of the life and times of bacteriophages via lectures or a textbook. Instead, the course offers an authentic chance for students to find new phages—and there are plenty. It’s estimated that the number of phages worldwide is 1031. In the course, students actually practice molecular biology, learn genetics by sequencing phage genomes, and chart phage evolution.
While there are a ton of phages in the world, not much is known about them. As a practical matter, the discovery and genetic profiling of new bacteriophages will shed light on what information their genes encode and what individual genes do, enabling a better understanding of phages’ role in the environment (they substantially impact carbon turnover rates) and their role in bacterial pathogenesis (how bacteria make us ill).
SEA-PHAGES also differentiates itself from other courses by introducing students to research methods and approaches, experiment design, and data interpretation in the context of a specific project (the phage hunt) rather than in generalities, allowing them to see exactly how abstract knowledge applies to authentic research.
The two-semester course begins with students digging in the dirt, then isolating phages from their soil samples. What happens next is essentially practicing microbiology as students grow a stock of phages to work with and examine them under an electron microscope, looking at their form and structure. Semester two is all about sequencing and studying their phage’s genome, using the tools of bioinformatics and computational biology. “They use complicated math to predict the location of genes, the organization of the genome, and to figure out what the genes do,” Hatfull says. “Then they examine how the genome of their phage is related to the other genomes out there.”
Other innovative science education courses, the mBio paper reports, work well on a smaller scale or target an already dedicated population of science students, but SEA-PHAGES has been successful because its curriculum is transferrable to institutions of various sizes, does not require tremendous phage-related expertise on the part of instructors, and allows students a sense of ownership that lecture courses without “wet bench” research components can’t provide.
Most institutions offer freshman biology labs; the more traditional among them have a “cookbook” aspect—basically a series of exercises to be conducted step-by-step. Some such courses are inquiry-based, where students can do exercises in the context of asking and addressing specific scientific questions. SEA-PHAGES goes further, which is to fully involve authentic scientific discovery (i.e., research) specifically in phage discovery and genomics.
“They start by digging in the dirt,” Hatfull says, “then what SEA-PHAGES does is give students experience in and engagement in real science. The rationale is that most students know what a doctor is, what she does, what a cop does, what a teacher does, but they don’t know what a scientist does. What they do know, they know from TV and movies and they think, ‘I could never do that; I’m not a genius.’”
Welkin Pope, research assistant professor of biological sciences, has been teaching the Pitt SEA-PHAGES course for two years. Like Hatfull, she endorses the hands-on aspect. “The students get to experience so many different things. First in the wet lab with classic techniques like phage identification and bacterial growth, and then using molecular techniques like polymerase chain reaction for DNA amplification. Finally we move into bioinformatics—and all in the same project.”
Enthusiasm for the Pitt-developed SEA-PHAGES is palpable beyond the University of Pittsburgh as well. “The SEA-PHAGES program at William and Mary has been absolutely amazing in its success and in its impact. It has made students from diverse backgrounds jazzed about science and research, and has helped them perform better in all their science classes,” says Margaret Saha, Chancellor Professor of Biology at the College of William and Mary in Williamsburg, Va. “From their first week on campus, freshmen are actually doing science and, for the first time, feeling part of a larger community, thanks to Professor Hatfull and his team. SEA-PHAGES has truly been transformative, here at William and Mary and nationwide!”
Pitt sophomore Jonathan Lapin is one of the SEA-PHAGES students who embraced the course and has become an undergraduate teaching assistant. His phage is named Velveteen, as in the children’s book The Velveteen Rabbit. (Lapin is French for “rabbit.”) “It was so much different than other huge intro classes with 400 students. This is a class where you get personal guidance with not only your research but with your science career. You’re not doing a lab project that comes out of a manual. You’re going to run into some failures, but that contributes to the learning process and you get a sense of pride and accomplishment,” he says.
The study, “A Broadly Implementable Research Course in Phage Discovery and Genomics for First-Year Undergraduate Students,” was supported by the Howard Hughes Medical Institute in Chevy Chase, Md.
Other Stories From This Issue
On the Freedom Road

Follow a group of Pitt students on the Returning to the Roots of Civil Rights bus tour, a nine-day, 2,300-mile journey crisscrossing five states.
Day 1: The Awakening
Day 2: Deep Impressions
Day 3: Music, Montgomery, and More
Day 4: Looking Back, Looking Forward
Day 5: Learning to Remember
Day 6: The Mountaintop
Day 7: Slavery and Beyond
Day 8: Lessons to Bring Home
Day 9: Final Lessons

